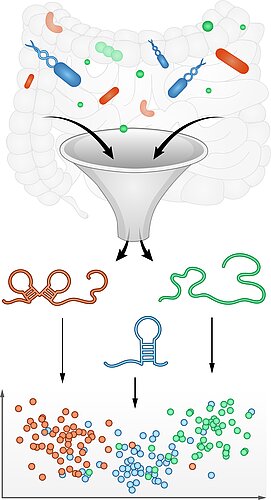
Discovery of functional RNAs in the microbiome
The major classes of noncoding RNA are known for very few bacterial species while the vast majority of microbes that matter to human health remains unexplored. lf we want to exploit RNA as a programmable antibiotic to treat bacterial infections and dysbiosis, we need to comprehensively understand the major RNA pathways and mechanisms in these species.
Over the years, we have pioneered a number of RNA deep sequencing-based methods such as differential RNA-seq (Sharma et al. 2010 Nature) or Dual RNA-seq (Westermann et al. 2016 Nature) to understand which RNAs are when expressed in the course of an infection. Moreover, Grad-seq (Smirnov et al. 2016 PNAS) now allows for an unbiased classification of potentially all transcripts of a cell based on their biochemical behavior.
We have set sail to chart the full cosmos of RNA functions in the human microbiome. This requires both, a strong component of method development to increase the resolution of Grad-seq and a biochemist's curiosity to elucidate the molecular m