Dr. Irene Beusch
Research
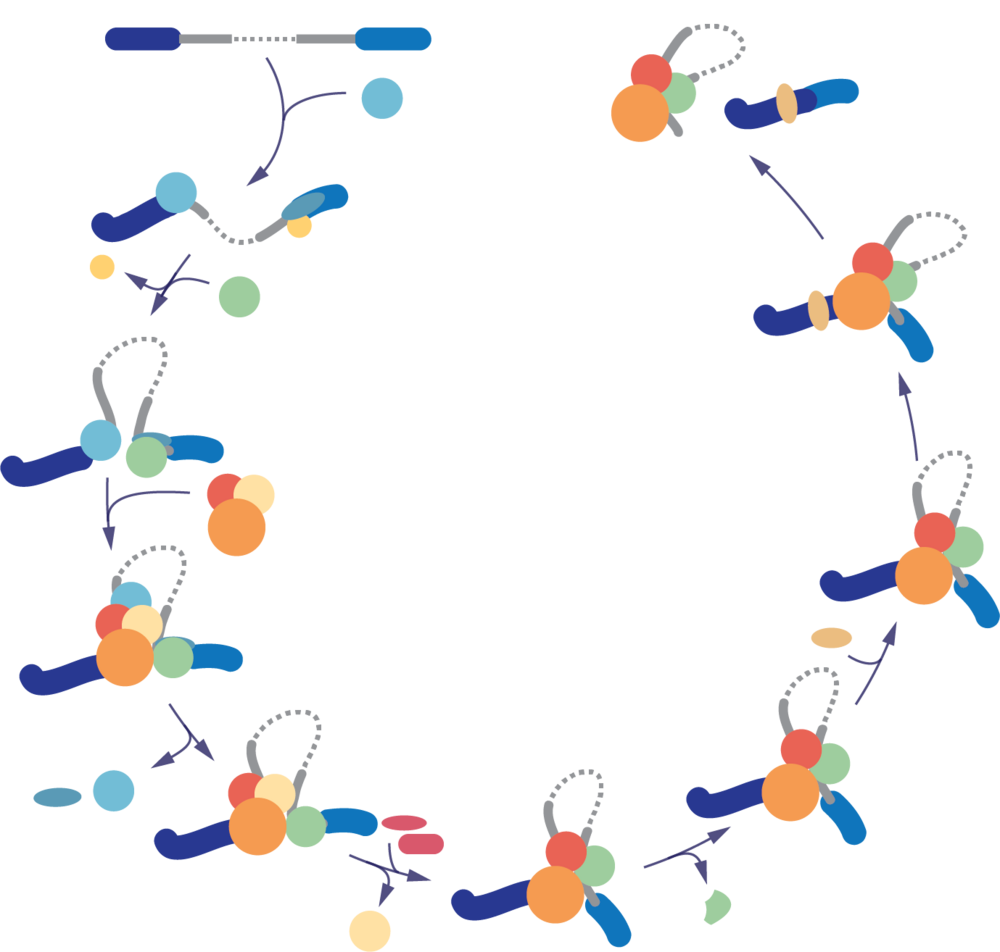
Within our research group we are interested in post-transcriptional processing with an emphasis on RNA splicing and the responsible cellular machinery, the spliceosome. Our research group’s goal is to gain a detailed mechanistic understanding of splicing and its regulation within the cell to not only understand fundamental eukaryotic biology but also human disease. In particular, we focus on understanding how splice site choice occurs and how this interplays with the assembly of the spliceosome. The spliceosome is a highly complex molecular machine, which will assemble de novo from over 150 proteins and 5 small nuclear RNAs for each reaction that it catalyses. What fascinates us is not only how this assembly is regulated but how the spliceosome can handle its very diverse substrate pool.
To do so, we use forward genetics in combination with a wide range of technologies including RNA-seq approaches as well as molecular biology methods and biochemistry.
Beusch I and Madhani HD (2024) Understanding the dynamic design of the spliceosome. Trends Biochem Sci 49(7):583-595 review
Beusch I, Rao B, Stude MK, Luhovska T, Šukytė V, Lei S, Oses-Prieto J, SeGraves E, Burlingame A, Jonas S, and Madhani HD (2023) Targeted high-throughput mutagenesis of the human spliceosome reveals its in vivo operating principles. Mol Cell 83(14):2578-2594
Sales-Lee J, Perry DS, Bowser BA, Diedrich JK, Rao B, Beusch I, Yates JR III, Roy SW, Madhani HD (2021) Coupling of spliceosome complexity to intron diversity. Curr. Biol. 31, 4898-4910.e4
Beusch I*, Barraud P*, Moursy A, Cléry A, Allain FHT (2017) Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. eLife 6, e25736
*equal contribution
Dr. Irene Beusch

Farimehr Etemadian

Famke Guder

Nandana Rajendran